Metabolism and Energy metabolism. (Determination
of pyruvic acid contents in urine).
Investigation of Krebs cycle functioning. (Determination of the muscles’ succinatedehydrogenase).
Energy releasing processes, ones that "generate"
energy, are termed exergonic reactions. Reactions that require energy to
initiate the reaction are known as endergonic reactions. All natural processes
tend to proceed in such a direction that the disorder or randomness of the
universe increases (the second law of thermodynamics).
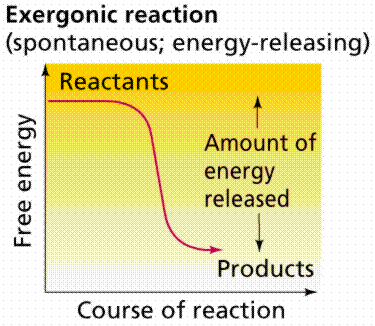
Time-energy graphs of an exergonic reaction (top)
and endergonic reaction (bottom). Images from Purves et al., Life: The
Science of Biology, 4th Edition, by Sinauer Associates (www.sinauer.com)
and WH Freeman (www.whfreeman.com),
used with permission.
Oxidation/Reduction
| Back to Top
Biochemical reactions in living organisms are
essentially energy transfers. Often they occur together, "linked", in
what are referred to as oxidation/reduction reactions. Reduction
is the gain of an electron. Sometimes we also have H ions along for the ride,
so reduction also becomes the gain of H. Oxidation
is the loss of an electron (or hydrogen). In oxidation/reduction reactions, one
chemical is oxidized, and its electrons are passed (like a hot potato) to
another (reduced, then) chemical. Such coupled reactions are referred to as
redox reactions. The metabolic processes glycolysis, Kreb's Cycle,
and Electron Transport Phosphorylation
involve the transfer of electrons (at varying energy states) by redox
reactions.
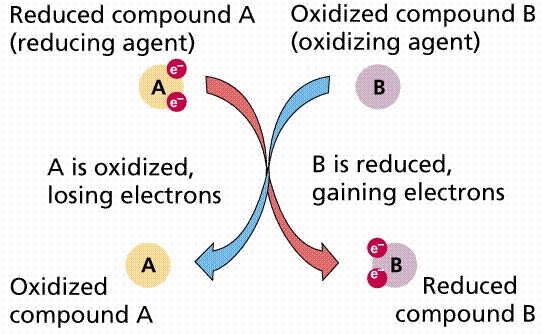
Passage of electrons from compound A to compound
B. When A loses its electrons it is oxidized; when B gains the electrons it is
reduced. Image from Purves et al., Life: The Science of Biology, 4th
Edition, by Sinauer Associates (www.sinauer.com)
and WH Freeman (www.whfreeman.com),
used with permission.

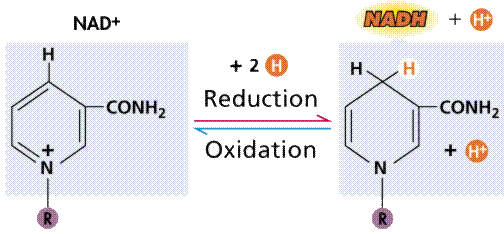
Oxidation/reduction via an intermediary (energy
carrier) compound, in this case NAD+. Images from Purves et al., Life:
The Science of Biology, 4th Edition, by Sinauer Associates (www.sinauer.com)
and WH Freeman (www.whfreeman.com),
used with permission.
Catabolism
and Anabolism | Back to Top
http://www.youtube.com/watch?v=MrOK_zWUzpM
Anabolism
is the total series of chemical reactions involved in synthesis of organic
compounds. Autotrophs must be able to manufacture (synthesize) all the organic compounds
they need. Heterotrophs
can obtain some of their compounds in their diet (along with their energy). For
example humans can synthesize 12 of the 20 amino acids, we must obtain the
other
Enzymes: Organic Catalysts | Back to Top
Enzymes
allow many chemical reactions to occur within the homeostasis
constraints of a living system. Enzymes function as organic catalysts. A
catalyst is a chemical involved in, but not changed by, a chemical reaction.
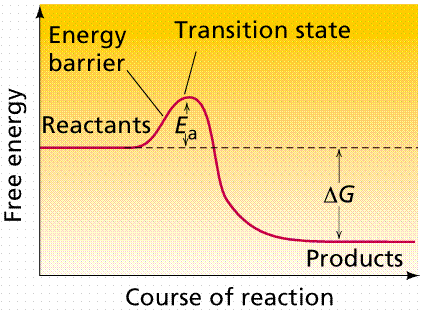
Many enzymes function by lowering the activation energy
of reactions. By bringing the reactants closer together, chemical bonds may be
weakened and reactions will proceed faster than without the catalyst.
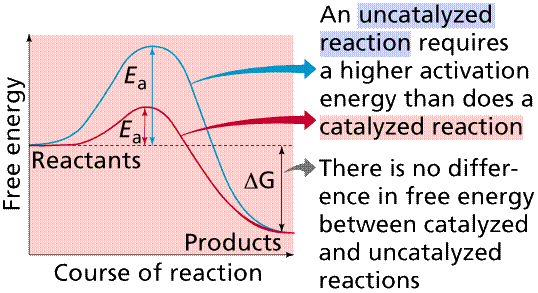
The use of enzymes can lower the activation energy of a reaction (Ea).
Image from Purves et al., Life: The Science of Biology, 4th Edition, by
Sinauer Associates (www.sinauer.com)
and WH Freeman (www.whfreeman.com),
used with permission.
Enzymes can act rapidly, as in the case of carbonic anhydrase (enzymes
typically end in the -ase suffix), which causes the chemicals to react 107
times faster than without the enzyme present. Carbonic anhydrase speeds up the
transfer of carbon dioxide from cells to the blood. There are over 2000 known
enzymes, each of which is involved with one specific chemical reaction. Enzymes
are substrate specific. The enzyme peptidase (which breaks peptide bonds in
proteins) will not work on starch (which is broken down by human-produced
amylase in the mouth).
Enzymes are proteins. The functioning of the enzyme is determined by the
shape of the protein. The arrangement of molecules on the enzyme produces an
area known as the active site within which the specific substrate(s) will
"fit". It recognizes, confines and orients the substrate in a
particular direction.
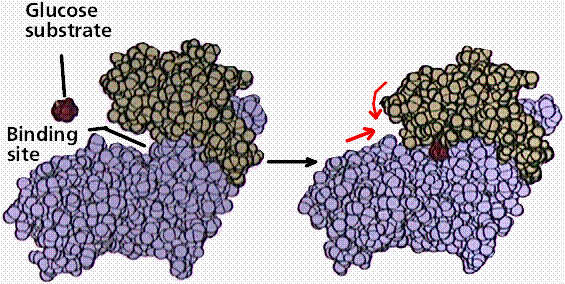
Space filling model of an enzyme working on glucose. Note the shape
change in the enzyme (indicated by the red arrows) after glucose has fit into
the binding or active site. Image from Purves et al., Life: The Science of
Biology, 4th Edition, by Sinauer Associates (www.sinauer.com)
and WH Freeman (www.whfreeman.com),
used with permission.
The induced fit hypothesis suggests that the binding of the substrate to
the enzyme alters the structure of the enzyme, placing some strain on the
substrate and further facilitating the reaction. Cofactors are nonproteins
essential for enzyme activity. Ions such as K+ and Ca+2
are cofactors. Coenzymes
are nonprotein organic molecules bound to enzymes near the active site. NAD (nicotinamide adenine dinucleotide).
A cartoonish view of the formation of an enzyme-substrate complex. Image
from Purves et al., Life: The Science of Biology, 4th Edition, by
Sinauer Associates (www.sinauer.com)
and WH Freeman (www.whfreeman.com),
used with permission.
Enzymatic pathways form as a result of the common occurrence of a series
of dependent chemical reactions. In one example, the end product depends on the
successful completion of five reactions, each mediated by a specific enzyme.
The enzymes in a series can be located adjacent to each other (in an organelle
or in the membrane of an organelle), thus speeding the reaction process. Also,
intermediate products tend not to accumulate, making the process more
efficient. By removing intermediates (and by inference end products) from the
reactive pathway, equilibrium (the tendency of reactions to reverse when
concentrations of the products build up to a certain level) effects are
minimized, since equilibrium is not attained, and so the reactions will proceed
in the "preferred" direction.
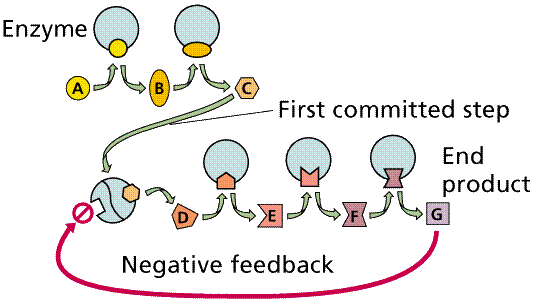
Negative feedback and a metabolic pathway. The production of the end
product (G) in sufficient quantity to fill the square feedback slot in the
enzyme will turn off this pathway between step C and D. Image from Purves et
al., Life: The Science of Biology, 4th Edition, by Sinauer Associates (www.sinauer.com)
and WH Freeman (www.whfreeman.com),
used with permission.
Temperature:
Increases in temperature will speed up the rate of nonenzyme mediated
reactions, and so temperature increase speeds up enzyme mediated reactions, but
only to a point. When heated too much, enzymes (since they are proteins
dependent on their shape) become denatured. When the temperature drops, the
enzyme regains its shape. Thermolabile enzymes, such as those responsible for
the color distribution in Siamese cats and color camouflage of the Arctic fox,
work better (or work at all) at lower temperatures.
Concentration of substrate and
product also control the rate of reaction, providing a
biofeedback mechanism.
Activation,
as in the case of chymotrypsin, protects a cell from the hazards or damage the
enzyme might cause.
Changes in pH
will also denature the enzyme by changing the shape of the enzyme. Enzymes are
also adapted to operate at a specific pH or pH range.
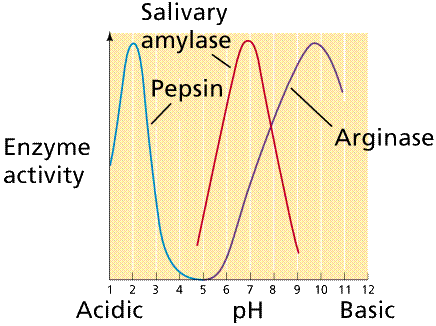
Plot of enzyme activity as a function of pH for
several enzymes. Note that each enzyme has a range of pH at which it is active
as well as an optimal pH at which it is most active. Image from Purves et al., Life:
The Science of Biology, 4th Edition, by Sinauer Associates (www.sinauer.com)
and WH Freeman (www.whfreeman.com),
used with permission.
Allosteric Interactions
may allow an enzyme to be temporarily inactivated. Binding of an allosteric
effector changes the shape of the enzyme, inactivating it while the effector is
still bound. Such a mechanism is commonly employed in feedback inhibition.
Often one of the products, either an end or near-end product act as an
allosteric effector, blocking or shunting the pathway.
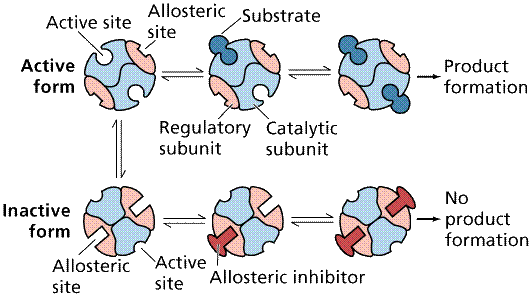
Action of an allosteric inhibitor as a negative control
on the action of an enzyme. Image from Purves et al., Life: The Science of
Biology, 4th Edition, by Sinauer Associates (www.sinauer.com)
and WH Freeman (www.whfreeman.com),
used with permission.
Competitive Inhibition
works by the competition of the regulatory compound and substrate for the
binding site. If enough regulatory compound molecules bind to enough enzymes,
the pathway is shut down or at least slowed down. PABA, a chemical essential to
a bacteria that infects animals, resembles a drug, sulfanilamide, that competes
with PABA, shutting down an essential bacterial (but not animal) pathway.
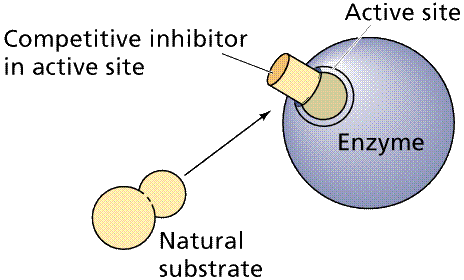
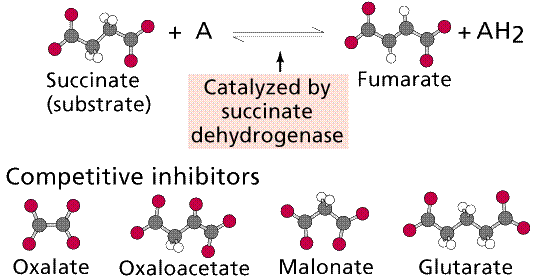
Top: general diagram showing competitor in the
active site normally occupied by the natural substrate; Bottom: specific case
of succinate dehydrogenase and its natural substrate (succinate) and
competitors (oxalate et al.). Images from Purves et al., Life: The Science
of Biology, 4th Edition, by Sinauer Associates (www.sinauer.com)
and WH Freeman (www.whfreeman.com),
used with permission.
Noncompetitive Inhibition
occurs when the inhibitory chemical, which does not have to resemble the
substrate, binds to the enzyme other than at the active site. Lead binds to SH
groups in this fashion. Irreversible Inhibition occurs when the chemical either
permanently binds to or massively denatures the enzyme so that the tertiary
structure cannot be restored. Nerve gas permanently blocks pathways involved in
nerve message transmission, resulting in death. Penicillin, the first of the
"wonder drug" antibiotics, permanently blocks the pathways certain
bacteria use to assemble their cell wall components.
Learning
Objectives | Back to Top
Reactions that show a net loss in energy are said
to be exergonic; reactions that show a net gain in energy are said to be endergonic.
Describe an example of each type of chemical reaction from everyday life.
What is meant by a reversible reaction? How might
this be significant to living systems?
What is the function of metabolic pathways
in cellular chemistry? Want more? Try Metabolic Pathways of Biochemistry.
What are enzymes?
Explain their importance.
Explain what happens when enzymes react with
substrates.
Links | Back to Top
Biology Project (U of A) Energy and Enzymes Problem Set
The
G6PD Deficiency Homepage 400 Million folks have this
problem, and it is enzymatic!
Enzyme Inhibition and Regulation (3/1/96)
From WSU's chemistry site.
MIT Hypertextbook Enzyme Chapter
Enzyme Reaction Tutorial
(U.C. Davis)
EC Enzyme Search the
EC enzyme databse, includes links to OMIM (Online Mendelain Inheritance in Man)
and SWISSPROT (Swiss Protein Database).
Interactive Cytochrome Oxidase
You will need the Chime plugin (available at this site), but it will be well
worth it. View either of the subunits of cytochrome oxidase as well as related
molecules. You can check buttons on the left frame to display selected portions
of the molecule, zoom in, and zoom out.
Metabolic
Pathways of Biochemistry Check out the metabolic
pathway of your choice in 2-D or 3-D (with the Chime plugin) models. is
provided than in most general biology textbooks, but the point is driven
home...a chemical reaction in one of these pathways needs its own enzyme.
Mitochondria (Mitochondria Under Microscope)
http://www.youtube.com/watch?v=XmrwRAytaMU&feature=related
Mitochondria
are thought to have evolved at least 1.8 billion years ago from primitive bacteria
which enjoyed such a symbiotic relationship with early eukaryotic cells.
Mitochondria still show some signs of their ancient origin. Mitochondrial
ribosome's are the 70S (bacterial) type, in contrast to the 80S ribosome's
found elsewhere in the cell. As in prokaryotes there is a very high proportion
of coding DNA, and an absence of repeats. Mitochondria genes are transcribed as
multigenic transcripts which are cleaved and polyadenylated to yield mature
mRNAs. Unlike their nuclear cousins, mitochondrial genes are small, generally
lacking introns, and the chromosomes are circular, conforming to the bacterial
pattern.
Discovery
Observed in 1850 by Kollicker in Muscle Cell.
Later Identified as the center for aerobicareobic respiration in the cell.
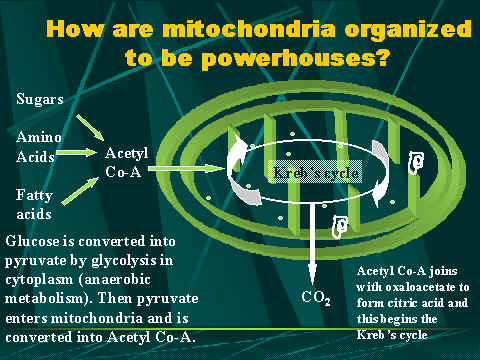
(Shows how a mitochondria is used in aerobic
respiration)
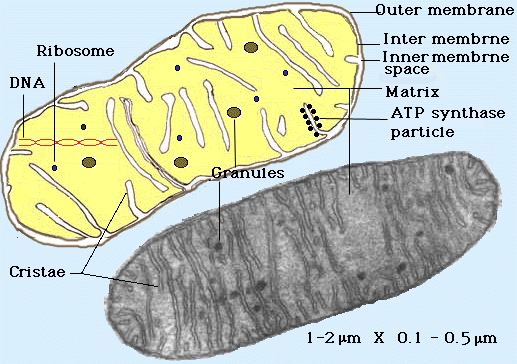
(Shows the inter and outer structure of a mitochondria) 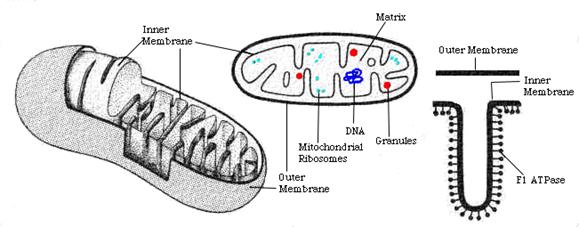
(Shows the outer and inter walls of a mitochondria in
great detail)
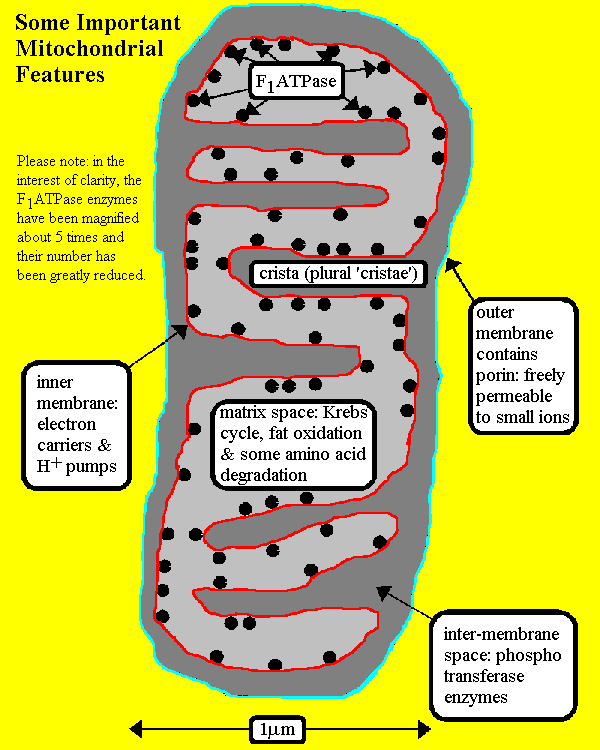
Mitochondria contain two membranes,
separated by a space. Both are the typical "unit membrane"
(railroad track) in structure. Inside the space enclosed by
the inner membrane is the matrix. This appears moderately dense and one
may find strands of DNA, ribosomes, or small granules in the matrix. The
mitochondria are able to code for part of their proteins with
these molecular tools. The above cartoon shows the diagram of the mitochondrial
membranes and the enclosed compartments.

(Shows the energy cycles that take place in the cell)
Cellular respiration is
the process of oxidizing food molecules, like glucose, to carbon dioxide and
water. The energy released is trapped in the form of ATP for use by all the
energy consuming activities of the cell. The process occurs in two
phases:
- glycolysis,
the breakdown of glucose to pyruvic acid
- the
complete oxidation of pyruvic acid to carbon dioxide and water
The energy conversion is as
follows:
C6H12O6 + 6O<2 -> 6CO2
+ 6H2O + energy (ATP)
Glycolysis:
There
are two important ways a cell can harvest energy from food: fermentation and cellular respiration.
Both start with the same first step: the process of glycolysis which is the breakdown or splitting
of glucose (6 carbons) into two 3-carbon molecules called pyruvic acid. The energy
from other sugars, such as fructose, is also harvested using this process.
Glycolysis is probably the oldest known way of producing ATP.
There is evidence that the process of glycolysis predates the existence of O2
in the Earth’s atmosphere and organelles in cells:
Glycolysis
does not need oxygen as part of any of its chemical reactions. It serves as a
first step in a variety of both aerobic and anaerobic energy-harvesting
reactions.
Glycolysis
happens in the cytoplasm of cells, not in some specialized organelle.
http://www.youtube.com/watch?v=p-lFJVOkFwk
Glycolysis is the one metabolic pathway found in
all living organisms.
Glucose
![]()
2
pyruvic acid molecules

+
4 H+ + energy stored in 2 ATP molecules

Fermentation:
In
fermentation these pyruvic acid molecules are turned into some “waste”
product, and a little bit of energy (only two ATP molecules per molecule of
glucose – actually four are produced in glycolysis, but two are used up) is
produced. Out of many possible types of fermentation processes, two of the most
common types are lactic acid fermentation and alcohol fermentation.

Pyruvic Acid + 2 H+
![]() or
or ![]()
Lactic
Acid  Ethanol
Ethanol 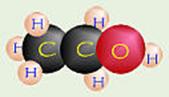
![]()
Carbon
Dioxide
Lactic
acid fermentation is done by some fungi, some bacteria like the Lactobacillus acidophilus. in
yogurt, and sometimes by our muscles. Normally our muscles do cellular
respiration like the rest of our bodies, using O2 supplied by our
lungs and blood. However, under greater exertion when the oxygen supplied by
the lungs and blood system can’t get there fast enough to keep up with the
muscles’ needs, our muscles can switch over and do lactic acid fermentation. In
the process of lactic acid fermentation, the 3-carbon pyruvic acid molecules
are turned into lactic acid. It is the presence of lactic acid
in yogurt that gives it its sour taste, and it is the presence of lactic acid
in our muscles “the morning after” that makes them so sore. Once our muscles
form lactic acid, they can’t do anything else with it, so until it is gradually
washed away by the blood stream and carried to the liver (which is able to get
rid of it), our over-exerted muscles feel stiff and sore even if they haven’t
been physically injured.
Alcohol
fermentation is done by yeast and some kinds of bacteria. The “waste” products
of this process are ethanol and carbon dioxide (CO2).
Humans have long taken advantage of this process in making bread, beer, and
wine. In bread making, it is the CO2 which forms and is trapped
between the gluten (a long protein in wheat) molecules that causes the bread to
rise, and the ethanol (often abbreviated as EtOH – do you remember how to draw
it?) evaporating that gives it its wonderful smell while baking. The effects of
the ethanol in beer and wine are something with which many college students are
familiar (sometimes too familiar?), and it is the CO2 produced by
the process of fermentation that makes these beverages effervescent.
Dr. Fankhauser has a number of fermentation-related recipes online,
complete with photographs:


His main cheese page
A recipe for cheese using one gallon of milk
A recipe for cheese using five gallons of milk
Homemade yogurt
Homemade buttermilk
Homemade root beer
Homemade ginger ale
A recipe for whole wheat bread
General information on milk-fermenting bacteria
Cellular Respiration:
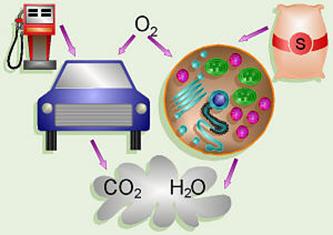
An analogy can be drawn between the process of cellular respiration in our
cells and a car. The mitochondria are the engines of our cells where sugar is
burned for fuel and the exhaust is CO2 and H2O. Note that
in a car that burned fuel perfectly, the only exhaust should theoretically be
CO2 and H2O also.
There
are three steps in the process of cellular respiration: glycolysis, the Krebs cycle, and the electron transport chain.
In contrast
to fermentation, in the process of cellular respiration, the pyruvic
acid molecules are broken down completely to CO2 and more energy
released. Note that three molecules of O2 must react with each
molecule of pyruvic acid to form the three carbon dioxide molecules, and three
molecules of water are also formed to “use up” the hydrogens. As mentioned
above, in glycolysis, a total of four molecules of ATP are produced, but two
are used up in other steps in the process. Additional ATP is produced during
the Krebs Cycle and the Electron Transport Chain, resulting in a grand total of
40 ATP molecules produced from the breakdown of one molecule of glucose via
cellular respiration. Since two of those are used up during glycolysis, in
prokaryotes a net total of 38 molecules of ATP are produced by cellular
respiration. Most prokaryotes have very simple cells which lack several types
of organelles present in eukaryotes, and therefore the Krebs Cycle and the
Electron Transport Chain occur in the cytoplasm and/or using chemicals embedded
in the cell membrane. In contrast, eukaryotes have more complex cells with more
specialized organelles to perform given functions. In eukaryotes, the Krebs
Cycle and Electron Transport Chain occur within the mitochondria, and thus the
pyruvic acid resulting from glycolysis must be sent into the mitochondria for
these reactions to occur. However, to move one molecule of pyruvic acid (remember
each molecule of glucose turns into two pyruvic acid molecules) from the
cytoplasm into a mitochondrion “costs” the cell one molecule of ATP (therefore
two ATPs for a whole glucose), thus a net total of 36 ATP molecules per
molecule of glucose is produced in eukaryotes as compared to only two in
fermentation. The overall reaction for cellular respiration is C6H12O6 + 6O2 ![]() 6CO2 + 6H2O
(+ energy for the cell to use for other things).
6CO2 + 6H2O
(+ energy for the cell to use for other things).
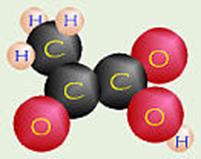
Pyruvic Acid + 2 H+
+
3 O2
![]()
![]()
![]()
![]()
3 Carbon Dioxide + 3 H2O+ 34 ATP
In glycolysis and the Krebs cycle, there are also
a number of electrons released as the glucose molecule is broken down. The cell
must deal with these electrons in some way, so they are stored by the cell by
forming a compound called NADH
by the chemical reaction, NAD+ + H+ + 2e– ![]() NADH.
This NADH is used to carry the electrons to the electron transport chain,
where more energy is harvested from them.
NADH.
This NADH is used to carry the electrons to the electron transport chain,
where more energy is harvested from them.
In
eukaryotes, the pyruvic acid from glycolysis must be transferred into the
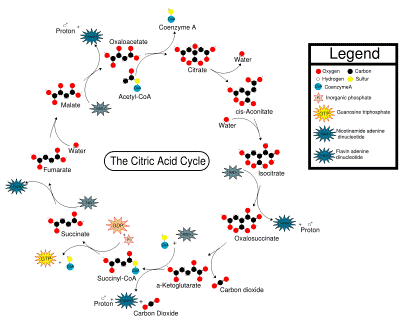
mitochondria to be sent through the Krebs cycle, also known as the citric acid cycle,
at a “cost” of one ATP per molecule of pyruvic acid. In this cycle, discovered
by Hans Krebs, the pyruvic acid molecules are converted to CO2, and
two more ATP molecules are produced per molecule of glucose. First, each
3-carbon pyruvic acid molecule has a CO2 broken off and the other
two carbons are transferred to a molecule called acetyl coenzyme A,
while a molecule of NADH is formed from NAD+ for each pyruvic acid
(= 2 for the whole glucose). These acetyl CoA molecules are put into the actual
cycle, and after the coenzyme A part is released, eventually each 2-carbon
piece is broken apart into two molecules of CO2. In the process, for
each acetyl CoA that goes into the cycle, three molecules of NADH, one molecule
of FADH2, and one molecule of ATP are formed (= 6 NADH, 2 FADH2,
and 2 ATP per whole glucose).
The electron transport chain is a system of electron
carriers embedded into the inner membrane of a mitochondrion.
As electrons are passed from one compound to the next in the chain, their
energy is harvested and stored by forming ATP. For each molecule of NADH which
puts its two electrons in, approximately three molecules of ATP are formed, and
for each molecule of FADH2, about two molecules of ATP are formed.
Many of the compounds that make up the electron
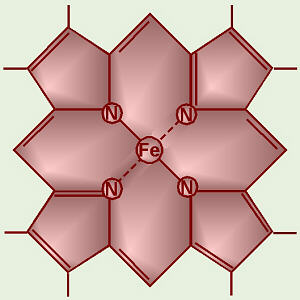
transport chain belong to a special group of chemicals called cytochromes. The central structure of a
cytochrome is a porphyrin ring
like chlorophyll but with iron in the center (chlorophyll has magnesium). A
porphyrin with iron in the center is called a heme group,
and these are also found in hemoglobin in our blood.
At
the last step in the electron transport chain, the “used up” electrons, along
with some “spare” hydrogen ions are combined with O2 (we finally got
around to the O2) to form water as a waste product: 4e- + 4H+ + O2 ![]() 2H2O.
2H2O.
Click on the heme group to see how to draw one.
Click
the picture to re-start or press [ESC] to stop. You may also “write” on the
picture. Unfortunately, Corel only has a Plug-In
for Win 95/NT, so this won’t work with Win 3.1 or Mac.
Many
of the enzymes in the cells of organisms need other helpers to function. These
non-protein enzyme helpers are called cofactors and can
include substances like iron, zinc, or copper. If a cofactor is an organic
molecule, it then is called a coenzyme. Many of the
vitamins
needed by our bodies are used as coenzymes to help our enzymes to do their
jobs. Vitamin B1 (thiamine) is a
coenzyme used in removing CO2 from various organic compounds. B2
(riboflavin) is a component of FAD (or FADH2),
one of the chemicals used to transport electrons from the Krebs cycle to the
electron transport chain. Vitamin B3 (niacin)
is a component of NAD+ (or NADH) which is the major
transporter of electrons from glycolysis and the Krebs cycle to the electron
transport chain. Without enough of these B vitamins, our ability to get the
energy out of our food would come to a grinding halt! B6 (pyridoxine), B12 (cobalamin), pantothenic acid, folic acid, and biotin
are all other B vitamins which serve as coenzymes at various points in
metabolizing our food. Interestingly, B12 has cobalt in it, a
mineral which we need in only very minute quantities, but whose absence can
cause symptoms of deficiency.
My
mother once had a friend who had porphyria, a dominant
genetic disorder in which the person’s body cannot properly make porphyrin
rings. This would, thus, affect the person’s ability to make both hemoglobin to
carry oxygen in the blood and cytochromes for the electron transport chain.
This woman’s symptoms were quite variable. At times, she would appear nearly
normal while on other occasions she would have to be hospitalized for temporary
paralysis of part of her body or other symptoms. There were a number of foods
and drugs she had to avoid because they would trigger or worsen her symptoms.
She frequently was in a lot of pain. Because porphyria is a dominant genetic
disorder, there was a 50% chance this woman’s daughter would also have
porphyria. Thus after the woman was diagnosed with porphyria, a number of tests
were also run on the girl, and she was more carefully monitored as she grew up.
My mother eventually lost contact with them, so I never heard the end of the
story.
Because
there are a number of enzymes and steps involved in forming porphyrin rings,
there are a number of possible points in the process where genetic defects
could occur. The Merck Manual says there are eight steps in the process of
making porphyrin rings, with genetic abnormalities possible in seven of the
eight enzymes.
Several
years ago, Dr. Fankhauser mentioned to me that he heard somewhere that an
“average”
First,
let’s assume that person eats an “average” dietary intake of 2500 KCal of food
energy (a number listed on the side of many food packages and a reasonable
amount that such a person might consume).
However,
just out of curiosity, let’s assume that all (100%) of that is glucose (In real
life, that would be a terrible idea! We need all the other nutrients that we
get from eating a variety of foods.). Since carbohydrates store about 4 KCal of
energy per gram, that would mean that 2500 KCal of glucose would be equivalent
to
Also,
just for the sake of argument, let’s assume that 100% of the ingested glucose
is burned for fuel, and that the process is 100% efficient so there is no waste
(in real life, our bodies would never use all 100% for fuel – some gets used to
build other chemicals, and just like the fuel efficiency in our automobiles,
the process is never 100% efficient.). Since, as was mentioned above,
eukaryotes make about 36 moles of ATP from every mole of glucose, then those
3.74 moles of glucose would be equivalent to 125 moles of ATP.
The
molecular weight of
ATP is 507 g/m, so that would be
Thus,
if it was really possible to meet all of those background assumptions and a
As
another example:
suppose
a person would consume one 12-oz. can of soft drink,
most
types of soft drink contain about 41 to
suppose
all of that sugar would be glucose,
suppose
the person’s body burns all of that sugar for fuel and does not store any of it
as fat or use any of it in other ways, and
suppose
the process of cellular respiration is 100% efficient and the sugar is
completely oxidized to CO2 and H2O.
Then:
since
the molecular weight of glucose is 180g/m, the
since
cellular respiration produces
since
the MW of ATP is 507 g/m, that would be equivalent
to
Recently I received an e-mail
message from a student who asked how long the whole process takes. While I have
never seen any information on that in print, a rough approximation can also be
calculated from the above statistic:
If, as mentioned above, an
“average”
40 kg/24 hr × 1
hr/60 min ×
1000 g/kg = about 27.8 g/min.
Since the molecular weight of
ATP is 507 g/m, then
that 27.8 g/min × 1
m/507 g = 0.0548 m/min.
Avagadro’s number says that
there are always 6.02 x 1023 molecules/mole,
so 0.0548 m/min ×
6.02 x 1023 molecules/mole = 3.30 x 1022 molecules/min.
or, since there are 60
sec/min, then that’s
3.30 x 1022 molecules/min × 1min/60 sec = 5.50 x 1020
molecules/sec made by a
so that would be equivalent to
5.50 x 1020 molecules/sec ÷
or × 1kg/1000 g =
7.85 ×
1015 molecules/sec/g of body
or × 1g/1000 mg =
7.85 ×
1012 molecules/sec/mg of body
or × 1mg/1000 µg
= 7.85 ×
109 molecules/sec/µg of body.
Krebs Cycle
http://www.youtube.com/watch?v=A1DjTM1qnPM&feature=related
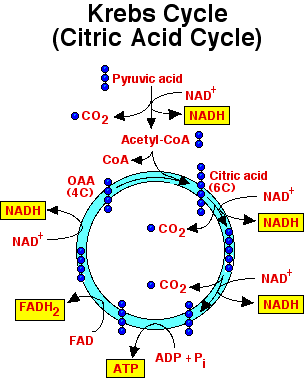
The pyruvate molecules produced during glycolysis contains a lot of
energy in the bonds between their molecules. In order to use that energy, the
cell must convert it into the form of ATP. To do so, pyruvate molecules are
processed through the Kreb Cycle, also known as the citric acid cycle.
http://www.youtube.com/watch?v=7gR4s8ool1Y
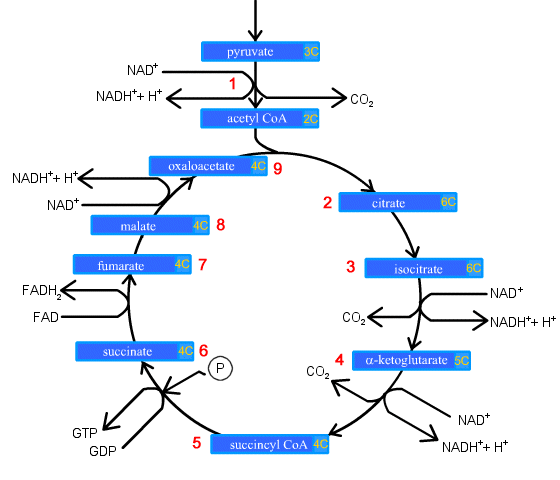
(Kerbs Cycle as a drawing)
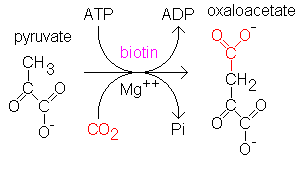
1. Prior to entering the Krebs Cycle, pyruvate must be converted into
acetyl CoA. This is achieved by removing a CO2 molecule from
pyruvate and then removing an electron to reduce an NAD+ into NADH.
An enzyme called coenzyme A is combined with the remaini ow:
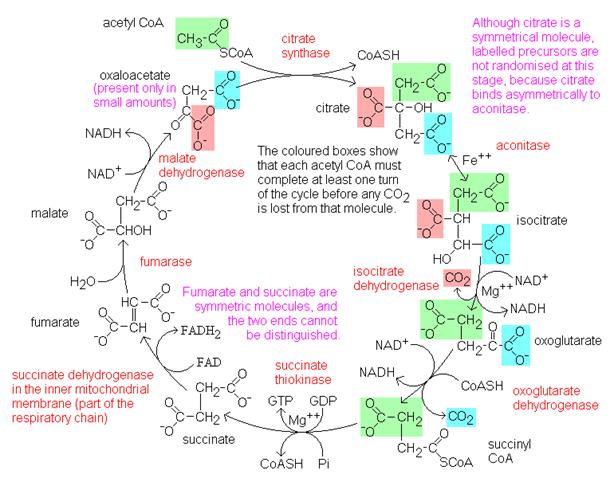
2. Citrate is formed when the acetyl group from acetyl CoA combines with
oxaloacetate from the previous Krebs cycle..
3. Citrate is converted into its isomer isocitrate..
4. Isocitrate is oxidized to form the 5-carbon α-ketoglutarate. This step releases one molecule of CO2
and reduces NAD+ to NADH2+.
5. The α-ketoglutarate is oxidized to
succinyl CoA, yielding CO2 and NADH2+.
6. Succinyl CoA releases coenzyme A and phosphorylates ADP into ATP.
7. Succinate is oxidized to fumarate, converting FAD to FADH2.
8. Fumarate is hydrolized to form malate.
9. Malate is oxidized to oxaloacetate, reducing NAD+ to NADH2+.
We are now back at the beginning of the Krebs Cycle. Because glycolysis
produces two pyruvate molecules from one glucose, each glucose is processes
through the kreb cycle twice. For each molecule of glucose, six NADH2+,
two FADH2, and two ATP.
These two pictures show the Electron Transport Chain
The schematic diagram above illustrates a
mitochondrion. In the animation, watch as NADH transfers H+ ions and
electrons into the electron transport system.
Key points:
1. Protons are translocated across the membrane, from the
matrix to the intermembrane space
2. Electrons are transported along the membrane, through
a series of protein carriers
3. Oxygen is the terminal electron acceptor, combining
with electrons and H+ ions to produce water
4. As NADH delivers more H+ and electrons into
the ETS, the proton gradient increases, with H+ building up outside
the inner mitochondrial membrane, and
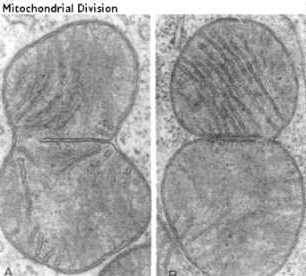
Mitochondria replicate like bacterial cells. When they get too
large they undergo fission. This involves a furrowing of the inner and
the outer membrane as if someone was pinching the mitochondria. The two
daughter mitochondria split. The Mitochondria must first replicate their
DNA.
Citric acid cycle
From Wikipedia, the free encyclopedia
Jump to:
navigation,
search
Overview
of the citric acid cycle
The citric
acid cycle (also known as the tricarboxylic acid cycle, the TCA
cycle, or the Krebs cycle, after Hans
Adolf Krebs who identified the cycle) is a series of chemical
reactions of central importance in all living cells
that use oxygen
as part of cellular respiration. In aerobic
organisms, the citric acid cycle is part of a metabolic
pathway involved in the chemical conversion of carbohydrates,
fats and proteins into carbon
dioxide and water
to generate a form of usable energy. It is the third of four metabolic pathways
that are involved in carbohydrate catabolism
and ATP production, the other three being glycolysis
and pyruvate oxidation before it, and respiratory
chain after it.
The
citric acid cycle also provides precursors for many compounds such as certain amino acids,
and some of its reactions are therefore important even in cells performing fermentation.
A
simplified view of the process
- The citric acid cycle begins with Acetyl-CoA
transferring its two-carbon acetyl group to the four-carbon acceptor compound,
oxaloacetate, forming citrate, a six-carbon compound.
- The citrate then goes through a
series of chemical
transformations, losing first one, then a second carboxyl
group as CO2.
- Most of the energy made available
by the oxidative steps of the cycle is transferred as energy-rich electrons
to NAD+, forming NADH. For each acetyl group that enters the citric acid cycle, three
molecules of NADH
are produced.
- Electrons
are also transferred to the electron acceptor FAD, forming FADH2.
- At the end of each cycle, the four-carbon
oxaloacetate has been regenerated, and the cycle continues. Products of
the first turn of the cycle are one GTP, three NADH, one FADH2,
and two CO2.
- Because two acetyl-CoA
molecules
are produced from each glucose molecule, two cycles are required per glucose molecule.
- At the end of all cycles, the
products are two GTP, six NADH, two FADH2,
four CO2.
Overview
The sum
of all reactions in the citric acid cycle is:
Acetyl-CoA + 3 NAD+ + FAD + GDP + Pi
+ 2 H2O → CoA-SH + 3 NADH + 3 H+ + FADH2
+ GTP + 2 CO2
(the
above reaction is equilibrated if Pi represents the H2PO4-
ion, GDP the GDP2- ion and GTP the GTP3- ion).
Two
carbons are oxidized
to CO2, and the energy from these reactions is stored in GTP, NADH and FADH2. NADH and
FADH2 are coenzymes (molecules that enable or enhance enzymes) that
store energy and are utilized in oxidative phosphorylation.
|
Step |
Substrate |
Enzyme |
Reaction type |
Products/ |
Comment |
|||
|
1 |
|
|
Acetyl CoA + |
CoA-SH |
|
|||
|
2 |
|
|
|
|
|
|||
|
3 |
|
|||||||
A simplified view of the process
- The citric acid cycle begins with Acetyl-CoA transferring its two-carbon acetyl group to the four-carbon
acceptor compound, oxaloacetate, forming citrate, a six-carbon compound.
- The
citrate then goes through a series of chemical transformations, losing
first one, then a second carboxyl group as CO2.
- Most of
the energy made available by the oxidative steps of the cycle is
transferred as energy-rich electrons to NAD+,
forming NADH. For each acetyl group that enters the
citric acid cycle, three molecules of NADH are produced.
- Electrons are also transferred to
the electron acceptor FAD, forming FADH2.
- At the end of each cycle, the
four-carbon oxaloacetate has been
regenerated, and the cycle continues. Products of the first turn of the
cycle are one GTP, three NADH, one FADH2, and
two CO2.
- Because
two acetyl-CoA molecules are produced from each glucose molecule, two cycles are required
per glucose molecule.
- At the
end of all cycles, the products are two GTP, six NADH, two FADH2, four
CO2.
Overall oxidation reactions of
Pyruvate and Glucose after the citric acid cycle
Pyruvic acid
+ 4 NAD+ + FAD + GDP + Pi + 2 H2O →
4 NADH + 4 H+ + FADH2 + GTP + 3 CO2
Regulation
Major metabolic pathways converging on the TCA cycle
MICROBIAL
GENETICS AND MICROBIAL METABOLISM
http://www.cat.cc.md.us/courses/bio141/lecguide/unit4/metabolism/cellresp/etsch.html
Gondar Design
Biology http://www.purchon.net/cells/mitochondria.htm
Journey into
the Cell: Mitochondria
http://biology.about.com/library/weekly/aa040600a.htm
Mitochondria:
Architecture dictates function
http://cellbio.utmb.edu/cellbio/mitoch1.htm
A Brief History of Mitochondria
http://www.a3243g.com/info_mitochondria_history.asp
Mitochondria:
Student Page http://www.fi.edu/qa97/biology/cells/cell9.html
The Virtual Cell Website http://personal.tmlp.com/Jimr57/textbook/chapter3/chapter3.htm
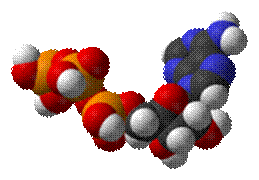
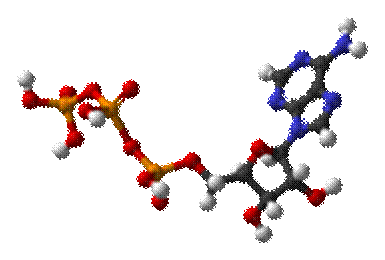
Adenosine
triphosphate
From Wikipedia,
the free encyclopedia
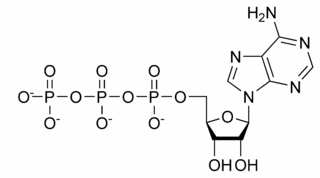
The structure of this molecule consists of a purine base (adenine) attached to the 1' carbon atom
of a pentose (ribose). Three phosphate groups are
attached at the 5' carbon atom of the pentose sugar. When ATP is used in DNA
synthesis, the ribose sugar is first converted to deoxyribose by ribonucleotide reductase. ATP was
discovered in 1929 by Karl Lohmann,[1] and was proposed to be the
main energy-transfer molecule in the cell by Fritz Albert Lipmann in 1941.[2] The Citric
Acid Cycle
Historical introduction
http://www.youtube.com/watch?v=hw5nWB0xN0Y&feature=related
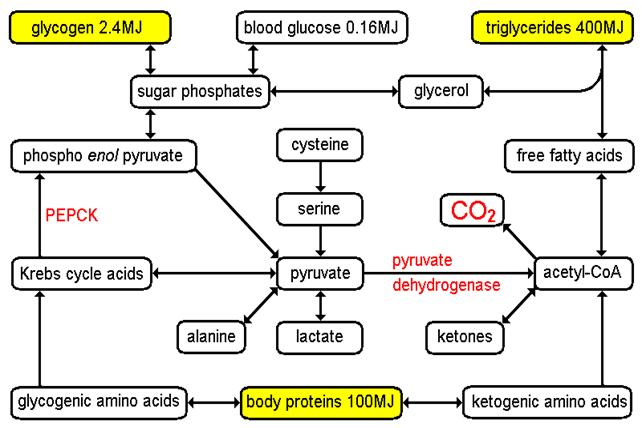
Energy stores and
inter-conversions in humans
(2) There
is no net
synthesis of amino acids under physiological conditions, but in the case of the non-essential
amino acids it may be possible to use transamination "to rob Peter to pay
Paul". aconitase (mitochondrial) citrate synthase fumarase (mitochondrial) isocitrate
dehydrogenase 3 (NAD, mitochondrial) malate
dehydrogenase (mitochondrial) methylmalonyl
CoA mutase methylmalonyl CoA racemase
oxoglutarate dehydrogenase propionyl CoA carboxylase pyruvate dehydrogenase succinate dehydrogenase succinate
thiokinase
This site
requires Netscape 4 or Internet Explorer 4 running JavaScript 1.2 or better.
Physical and
chemical properties
Ionization in
biological systems
Biosynthesis
[edit] Glycolysis
[edit] Citric acid cycle
http://www.youtube.com/watch?v=3OmMOcbTqE4&feature=related
Main
articles: Citric acid cycle and oxidative phosphorylation
Beta-oxidation
Anaerobic
respiration
Main article: anaerobic respiration
C6H12O6 ---> 2CH3CH(OH)COOH
+ 2 ATP
[edit] ATP replenishment
by nucleoside diphosphate kinases
ATP production during photosynthesis
[edit] ATP recycling
Regulation
of biosynthesis
which directly
implies this equation:
Functions
in cells
Extracellular signaling
ATP is also a signaling molecule. ATP, ADP, or
adenosine are recognized by purinergic receptors.
![\frac{cyt~c_{red}}{cyt~c_{ox}} = \left(\frac{[NADH]}{[NAD]^{+}}\right)^{\frac{1}{2}}\left(\frac{[ADP][P_{i}]}{[ATP]}\right)K_{eq}](Metabolism%20and%20Energy%20metabolism%20Investigation%20of%20Krebs%20cycle%20functioning..files/image058.gif)